Contents
Quick start
Simply enter the DNA variant you would like to analyse into the variant field, select one or multiple transcription factors and click on Analyse. If you do not know the location but have a wild-type and a variant sequence, you can still enter them by clicking on Enter sequences directly.
Interfaces
Click on an image to enlarge it.
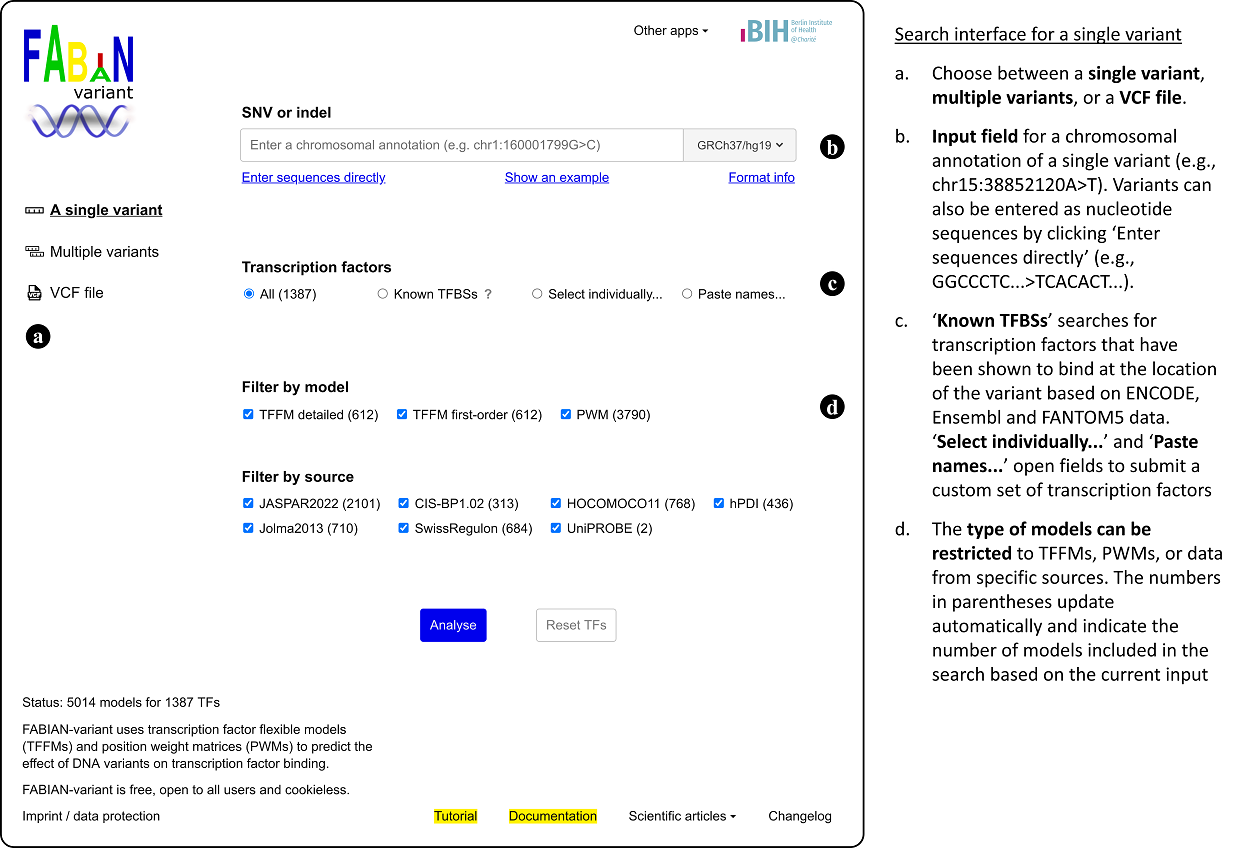
Variant input
FABIAN-variant supports five different input modes for variants. In each mode the supported formats can be displayed by clicking on the link "Format info" below the input field.
- Single variant: Chromosome, genomic position, reference base(s) and variant base(s) have to be entered in one of the following formats:
2:232526664T>C(default)2:232.526.664T>C(dot as thousands separator)chr2:g.232,526,664T>C(comma as thousands separator)2-232526664-T-C(gnomAD)2 232526664 . T C(VCF)2 232526664 T C(VCF without ID column)
- Single variant (sequences): The wild-type sequence and variant sequence may consist of letters ACGT. We suggest to use sequences of about 30 bases. For example:
- Wild-type sequence:
GGCCCTCACACTCTCCAACCTCATCTCCCTGGTGAGAGGCC - Variant sequence:
TCACACTCTCCAACCTCATCTCCCTGGTGAG
- Wild-type sequence:
- Multiple variants: The accepted formats are the same as for a single variant (see above). One variant has to be entered per line (separated by newline character). GRCh37/hg19 or GRCh38/hg38 must match the chromosomal annotation. For example:
17:19533822G>A
1:778570CTG>C
- Multiple variants (sequences): The only accepted format is a space-separated list of wild-type and variant sequences with one variant per line (separated by newline character). For example:
GGCCCTCACACTCTCCAA TCACACTCTCCAA
ATAAATTTTTTTT ATAAAGGGTTTTT
TCTTCTTCCAGCGGAGGCGGGATT TCTTCTTCCAGCGGACGCGGGATT
<WT> <MT>where each <WT> sequence and <MT> sequence may consist of letters ACGT. A space character is used to separate <WT> and <MT> and a newline character to separate two variants. We suggest to use
<WT> <MT>
<WT> <MT>
...and sequences of about 30 bases.
- VCF file:
Input files have to be in VCF format.
The uploaded VCF file may only contain data from one sample (support for multi-sample VCF files is limited). Please zip or gzip large VCF files before upload.
- The contents of a small sample VCF file (TinyExample38.vcf) are printed below:
3 190404905 . G A 116 . . GT:DP 0/1:154 1 1048791 . CAG C 116 . . GT:DP 0/1:154 21 31663857 . A G 116 . . GT:DP 0/1:154 - A sample whole genome sequencing VCF file can be found in the tutorial.
- Very low covered positions do not offer reliable data. Therefore, it is useful to exclude such variants from analysis.
FABIAN-variant offers the possibility to skip variants that are covered below a user-defined threshold.
To this end, adjust the number in the corresponding "Minimum coverage" text field in the VCF options.
If you do not want to exclude poorly covered variants or if your VCF does not have
DPvalues, fill in 0. The default minimum coverage is 10. - The GT values determine which alternative alleles in each VCF line are analyzed:
1for the first alternative allele and2for the second alternative allele (see VCF documentation for details). Diploid and haploid calls are supported. Alternatively, if you do not specify GT, all alternative alleles will be analyzed. - In general, when using a multi-sample VCF with FABIAN-variant, only the first sample is looked at. For example, if the GT value of the first sample for a particular variant is 0/0, this variant will be classified as "is_refseq" and skipped. Skipped variants are not analyzed and are included in a file called "skipped.txt" that can be downloaded from the results page. By selecting the option 'Ignore GT and analyze all ALT alleles', you can bypass the GT checks and allow analysis of all variants in a multi-sample VCF file.
- You may specify filter options to skip analysis of your variants that were also found in gnomAD, ExAC or the 1000Genomes Project (1000G). If you wish to exclude variants found 10 or more times (gnomAD or ExAc) or 4 or more times (1000G) in homozygous state but include all heterozygous variants, you can leave everything as it is (default setting). But you are free to change the number of cases that have to be present in either source in order to exclude variants from analysis.
- The contents of a small sample VCF file (TinyExample38.vcf) are printed below:
Transcription factors
FABIAN supports more than 5,000 different binding models for 1387 human transcription factors. The models were pooled from various publicly accessible data sources:
Many of these data sources were obtained from MotifDb, which is an annotated collection of PWM models. 1224 transcription factor flexible models (TFFMs) from JASPAR are included. For each transcription factor, FABIAN-variant combines the results of different models for a final prediction of the resulting binding affinity change.
The underlying data is available for download. It contains:
- Definitions of 1224 TFFMs
- Definitions of 3790 matrices
TFFM definitions were converted from XML to a flat file format to improve processing in FABIAN-variant.
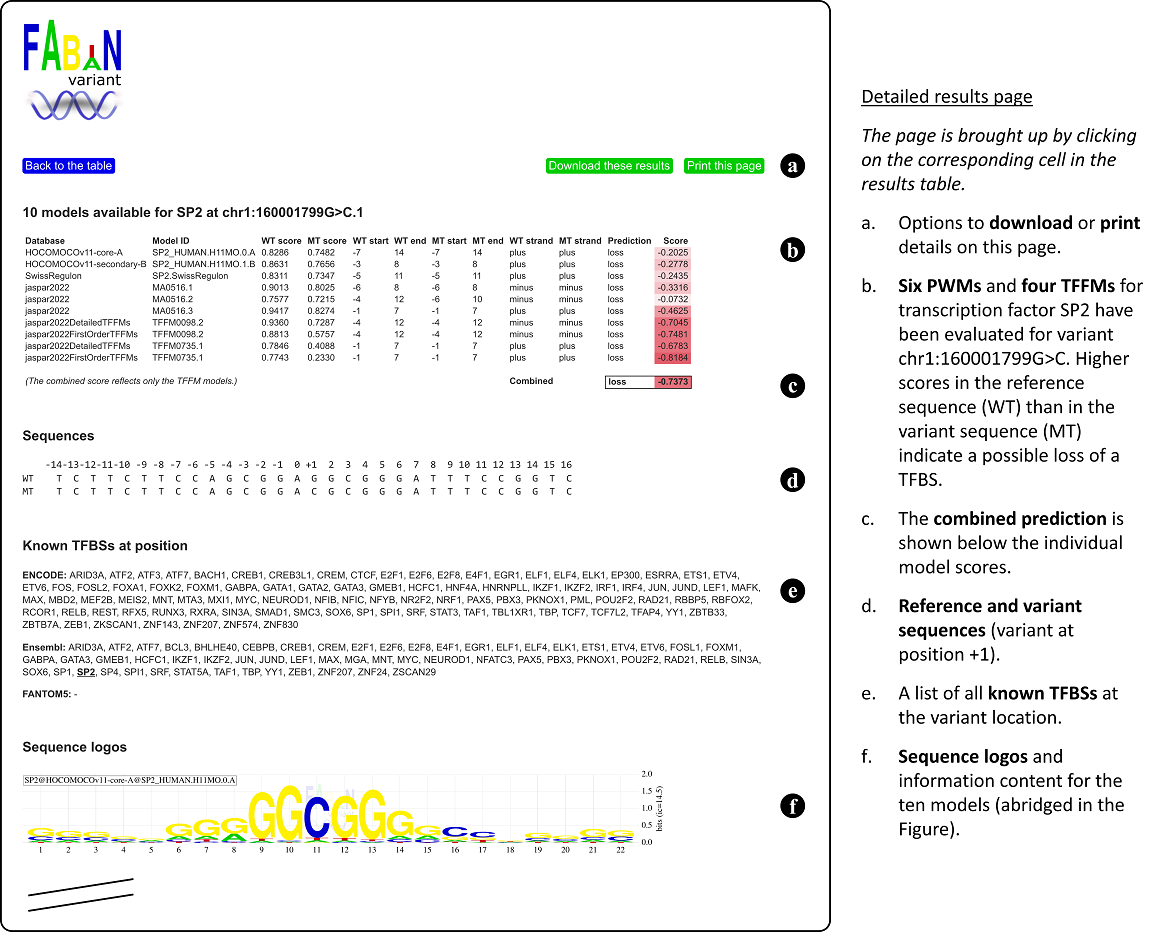
Known transcription factor binding sites
On the results page, FABIAN-variant highlights known binding sites for transcription factors by a black rectangle around the score. Genome locations of known binding sites were pooled from these sources:
Please note that this function is only available if you entered genomic positions. As the TFBS sites provided by ENCODE and Ensembl are several hundred bases long, there is not necessarily really a binding site for your TF at your exact position.
Evaluation of models
TFFMs and PWMs are evaluated in the window [-15,15] around the variant in both strands and in both the
reference sequence (WT)
and the mutated sequence (MT).
The highest score in the mutated sequence is compared with the highest score in the reference sequence.
A greater WT score indicates a weakened binding affinity, and a greater MT score indicates an increased binding affinity due to the variant.
For each model, FABIAN-variant generates a joint score S between -1 (likely TFBS loss) and +1 (likely TFBS gain),
with pseudocount α = 0.1 to avoid zero in the denominator. This link illustrates the function in an interactive plot for different values 0 ≤ WT ≤ 1 and 0 ≤ MT ≤ 1.
To obtain the combined prediction from multiple models, FABIAN-variant calculates the average of joint scores S of the individual models. If both TFFMs and PWMs are available, by default only the results from TFFMs are used for the combined prediction (this setting can be changed by unchecking "Options > Prefer TFFMs" on the results page).
The WT score, MT score, the joint score S per model and the combined score are shown on the results page. For example:

Evaluation of TFFMs
A C++ implementation of the forward-backward algorithm evaluates TFFMs. See this article to learn more about TFFMs:
Mathelier A, Wasserman WW. The next generation of transcription factor binding site prediction. PLoS computational biology. 2013 Sep 5;9(9):e1003214. https://doi.org/10.1371/journal.pcbi.1003214
There a two types of TFFMs: Detailed models and first-order models. Detailed models are always listed
as jaspar2022DetailedTFFMs and first-order models as jaspar2022FirstOrderTFFMs in the database field in the results table.
The model ID field starts with TFFM (e.g., TFFM0040.1).
Evaluation of PWMs
Position count matrices (PCMs) were converted to position weight matrices (PWMs) using the method described in:
Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. Journal of molecular biology. 1990 Apr 20;212(4):563-78. https://doi.org/10.1016/0022-2836(90)90223-9
A custom C++ implementation computes the scores.
In the results table, the database field for PWMs is one of the following:
jaspar2022,
cisbp_1.02,
HOCOMOCOv11-core-B,
HOCOMOCOv11-core-C,
HOCOMOCOv11-secondary-D,
HOCOMOCOv11-core-A,
HOCOMOCOv11-secondary-A,
HOCOMOCOv11-secondary-B,
HOCOMOCOv11-secondary-C,
hPDI,
jolma2013,
SwissRegulon,
UniPROBE.
Results page
The results table summarizes predictions from different models per variant and transcription factor on coloured scales for a possible loss (red) or gain (blue) of a TFBS. Deeper shades of the colour represent a higher loss or gain. Known TFBSs are displayed with a border around the cell.
Moving the mouse pointer over a coloured cell reveals the individual model scores. Clicking on the table cell shows the detailed results page.
Variants have the format chr1:778570CTG>C.1 or GGCCCTCAC>TCACACTCTCCAACCT*.1.
In both cases, .1 is simply the line number of the variant in the input. * indicates that some bases of
a long sequence are not displayed.
Clicking on a variants opens the corresponding location in the UCSC Genome Browser. Clicking on a transcription factor opens Ensembl.
The results table can be filtered and sorted in the browser using the checkboxes and radio buttons in the header of the page:
- Show effects
- TFBS loss: Display TFs with at least one predicted TFBS loss in the visible variants (combined score < 0)
- TFBS gain: Display TFs with at least one predicted TFBS gain in the visible variants (combined score > 0)
- Neutral: Display TFs with no predicted effect in the visible variants
- Sort/filter TFs
- Sort by name: TFs are shown in alphabetical order (default).
- Sort by loss: TFs are sorted by the combined loss scores over all visible variants.
- Sort by gain: TFs are sorted by the combined gain scores over all visible variants.
- With known BS: Only display TFs with at least one known TFBS in visible variants.
- Manual selection...: When activating the checkbox, all TF names are immediately highlighted grey (selected state). Click on a TF name to change the selection. Then use the <ENTER> key to keep all selected TFs or use the <BACKSPACE> key to hide all selected TFs.
- Filter variants
- In known TFBSs: Only show variants with at least one known binding sites of the visible TFs.
- Manual selection...: When activating the checkbox, all variants are immediately highlighted grey (selected state). Click on a variants to change the selection. Then use the <ENTER> key to keep all selected variants or use the <BACKSPACE> key to hide all selected variants.
- By position...: When activating the checkbox, an input field opens where you can enter chromosome and position. The following formats are accepted: chr1 (whole chromosome), chr1:155259323 (exact position), chr1:155259000-155260000 (range)
- Sort variants
- Input: Variants are shown in the order in which they were listed in the input (default).
- Position: Variants are sorted by chromosome and position.
- TFBS loss: Variants are sorted by the combined loss scores over all visible TFs.
- TFBS gain: Variants are sorted by the combined gain scores over all visible TFs.
- Options
- Prefer TFFMs: When this option is selected (default), the combined score is calculated using only TFFMs when both PWMs and TFFMs exist for the same transcription factor. When this option is not selected, both types of models are included in the combined score. The checkbox is automatically disabled when only one type of model was selected on the search input page.
- Columns view: By default, results of only a few variants are shown in multiple columns for better readability. The checkbox can be used to disable this feature.
- Reset view: This resets all filters back to their default state.
- Show log: This option displays or hides the computation log.
- Show links: By default, clicking on TF names in the results table opens the Ensembl gene summary and clicking on a variant name shows the UCSC genome browser. This checkbox hides the links.
Results retention
Results are kept available on the server for three days after the analysis is complete. After this time, they are automatically deleted. You can also manually delete your results by checking the "Options > Show log" checkbox on the top menu on the results page and clicking on the "delete" link. Deleting results also removes all information about your search parameters and uploaded variants from our servers. Deleted results cannot be restored.
Download format
The full download of all results has the following columns:
variant tf model_id database model_db wt_score mt_score start_wt end_wt start_mt end_mt strand_wt strand_mt prediction score
variant: The name of the variant has the formatchromosome : position REF ALT . variant_numbertf: Name of the transcription factormodel_id: ID of the model in the source databasedatabase: Source database of the modelwt_score,mt_score: The highest score in the reference and mutated sequencestart_wt,end_wt: Location with the highest score in the reference sequence relative to the variantstart_mt,end_mt: Location with the highest score in the mutated sequence relative to the variantstrand_wt,strand_mt: Strand of the location with the highest score in the reference and mutated sequenceprediction: Prediction of a gain or loss of TFBS, or NA if not prediction was possiblescore: Score of prediction between -1 (likely TFBS loss) and +1 (likely TFBS gain)
The summary download is similar to the results table and includes any filters and sorting options at the top of the results page.
Scores for a known TFBSs are marked with *.
Programmatic access
On Unix-based systems, you can use cURL to post variants to and receive results from FABIAN-variant. The general pattern is printed below.
printf "($(date +%T)) Submitting " && \
FABIANID=$( curl -sLD - -o /dev/null \
-F "mode=vcf" \
-F "filename=@TinyExample38.vcf" \
-F "genome=hg38" \
-F "tfs_filter=all" \
-F "models_filter=tffm_d" \
-F "models_filter=tffm_fo" \
-F "models_filter=pwm" \
-F "dbs_filter=jaspar2022" \
-F "dbs_filter=cisbp_1.02" \
-F "dbs_filter=HOCOMOCOv11" \
-F "dbs_filter=hPDI" \
-F "dbs_filter=jolma2013" \
-F "dbs_filter=SwissRegulon" \
-F "dbs_filter=UniPROBE" \
https://www.genecascade.org/fabian/analyse.cgi \
| grep -m 1 "Location: " | grep -o "\([0-9]\+_[0-9]\+\)" ) && \
i=1; until curl -sfo fabian.data_${FABIANID}.zip \
https://www.genecascade.org/temp/QE/FABIAN/${FABIANID}/fabian.data.zip; \
do printf "\r($(date +%T)) Waiting for $FABIANID"; \
[ $i == 30 ] && sleep $i || sleep $((i++)); done && \
printf "\r($(date +%T)) Saved file fabian.data_${FABIANID}.zip\n"Some parameters are specific depending on the mode and which transcription factors you are looking for. A few examples are listed below.
- Single variant:
-F "mode=single" \ -F "single_hgvs=1:160032009G>C" \ -F "genome=hg38" \ - Single variant (sequences):
-F "mode=single_seq" \ -F "single_wt=GGCCCTCACACTCTCCAACCTCATCTCCCTGGTGAGAGGCC" \ -F "single_mt=TCACACTCTCCAACCTCATCTCCCTGGTGAG" \ - Multiple variants:
-F "mode=batch" \ -F "batch_hgvs=17:19533822G>A 1:778570CTG>C" \ -F "genome=hg38" \ - Multiple variants (sequences):
-F "mode=batch_seq" \ -F "batch_wt_mt=GGCCCTCACACTCTCCAA TCACACTCTCCAA ATAAATTTTTTTT ATAAAGGGTTTTT" \ - VCF file:
Note that the path to the VCF file must be prefixed with "@".-F "mode=vcf" \ -F "filename=@TinyExample38.vcf" \ -F "genome=hg38" \
- Search for individual transcription factors:
-F "tfs_filter=names" \ -F "tfs_filter_names_tb=SP1 SP2 SP3 SP4" \ - Get notified via email when results are available:
-F "email=your@email.edu" \
If the request is correct, cURL polls our server until results are available, which are then saved under a project-specific name (e.g., fabian.data_1650751034_19489.zip).
Please note that your request may wait indefinitely in case of an error. You can always check the status at the project-specific URL (e.g., https://www.genecascade.org/fabian/1650751034_19489)
Please do not run more than three automated requests at the same time! If you require more processing slots, please send us a short email with details of your request.
Team
FABIAN has been developed at Berlin Institute of Health (BIH) by
- Robin Steinhaus
- Peter N. Robinson
- Dominik Seelow
FABIAN is an update of the ePOSSUM software.
Contact
If you have suggestions about this software, please do not hesitate to email robin.steinhaus (at) bih-charite.de. If you discover a bug, please submit a ticket via email using this link.